Package Exports
- jupyter-cytoscape
This package does not declare an exports field, so the exports above have been automatically detected and optimized by JSPM instead. If any package subpath is missing, it is recommended to post an issue to the original package (jupyter-cytoscape) to support the "exports" field. If that is not possible, create a JSPM override to customize the exports field for this package.
Readme
ipycytoscape
Python implementation of the graph visualization tool Cytoscape.
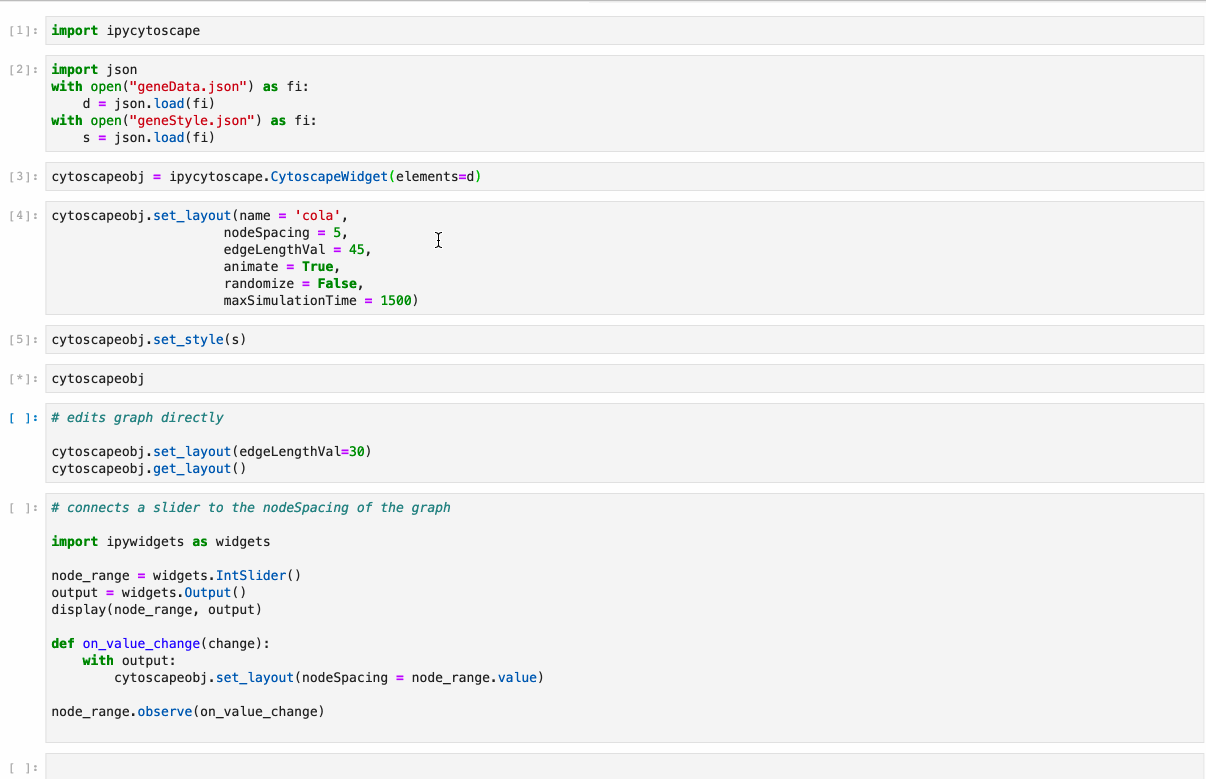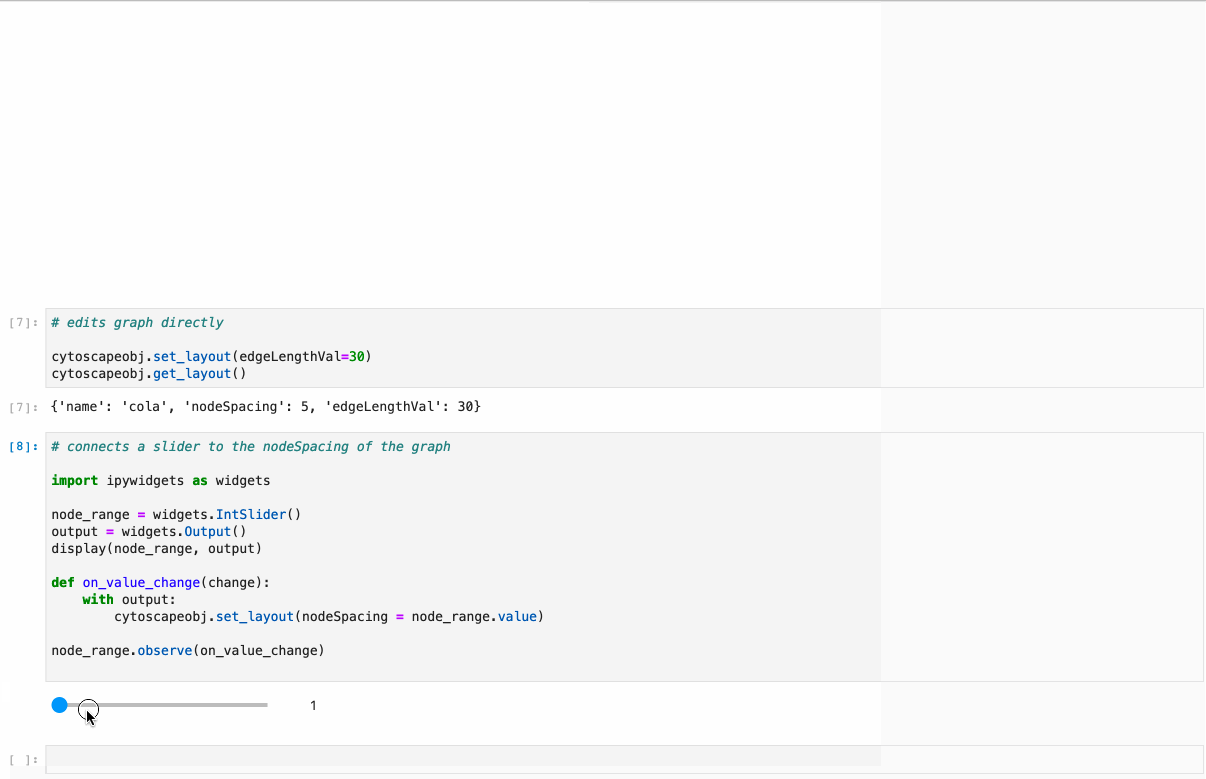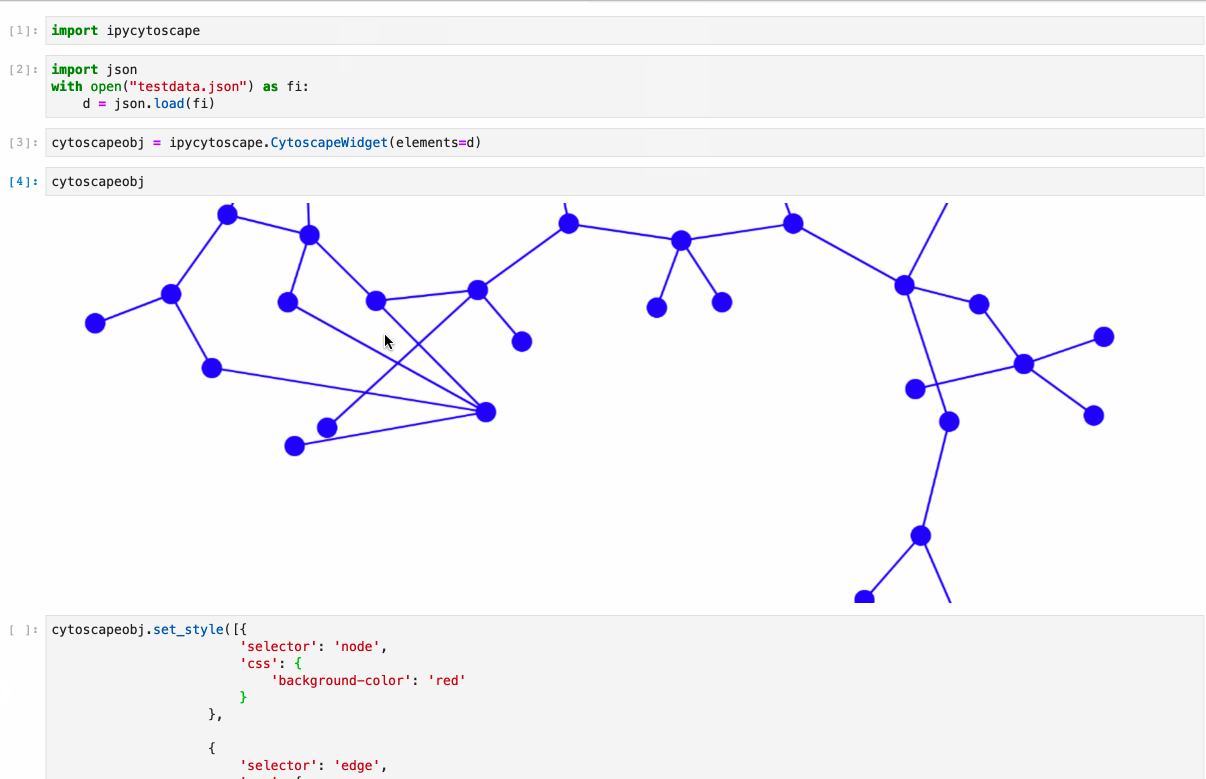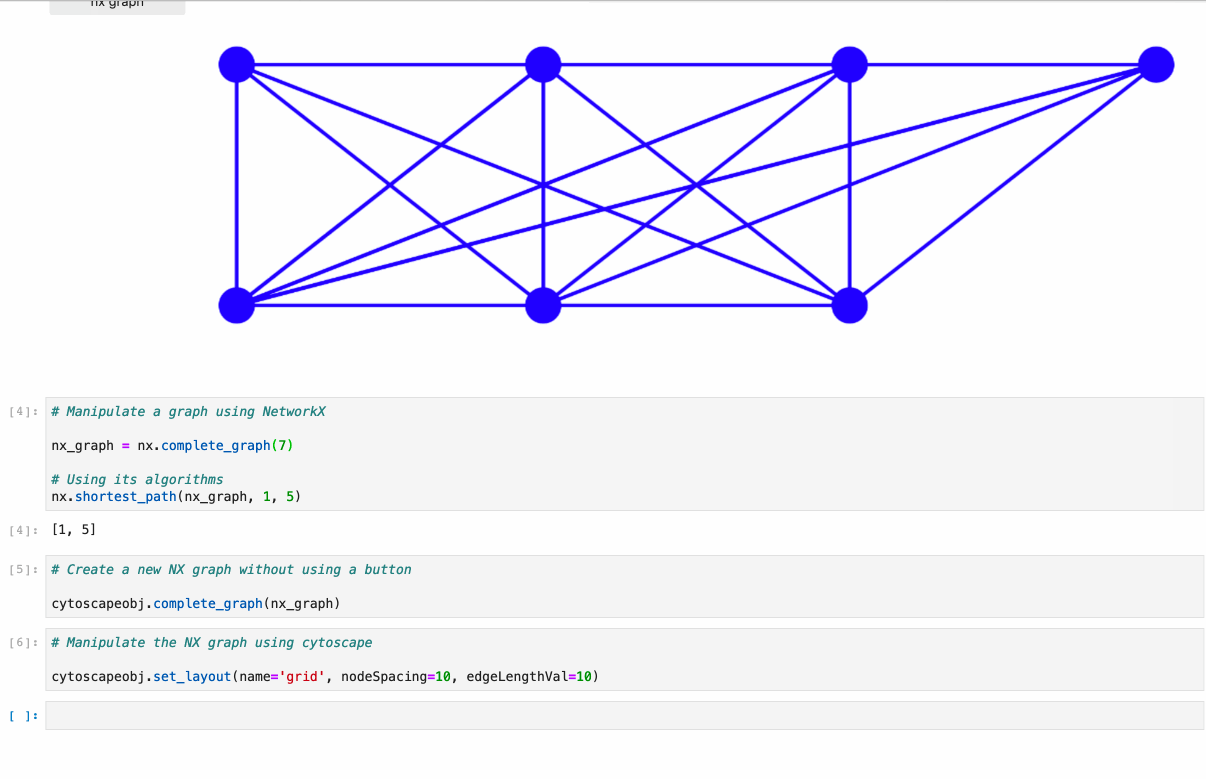
Offers full support to NetworkX lib. Just follow the example under /examples/Test NetworkX methods.ipynb.
Installation
With conda: (recommended)
conda install -c conda-forge ipycytoscapeWith pip:
pip install ipycytoscapeOr if you use jupyterlab:
pip install ipycytoscape
jupyter labextension install @jupyter-widgets/jupyterlab-manager
jupyter labextension install jupyter-cytoscape@0.1.2If you are using Jupyter Notebook 5.2 or earlier, you may also need to enable the nbextension:
jupyter nbextension enable --py [--sys-prefix|--user|--system] ipycytoscapeFor a development installation: (requires npm)
$ git clone https://github.com/QuantStack/ipycytoscape.git
$ cd ipytree
$ pip install -e .
$ jupyter nbextension install --py --symlink --sys-prefix ipycytoscape
$ jupyter nbextension enable --py --sys-prefix ipycytoscape
$ jupyter labextension install @jupyter-widgets/jupyterlab-manager
$ jupyter labextension install jsLicense
We use a shared copyright model that enables all contributors to maintain the copyright on their contributions.
This software is licensed under the BSD-3-Clause license. See the LICENSE file for details.